Deep MIB - Directories and Preprocessing Tab
Back to MIB | User interface | DeepMIB
Configuration of directories and preprocessing settings for deep learning segmentation in Microscopy Image Browser.
Overview
The Directories and Preprocessing tab in Deep MIB allows users to specify directories containing images for training and prediction, along with parameters for image loading and preprocessing.
Widgets and settings
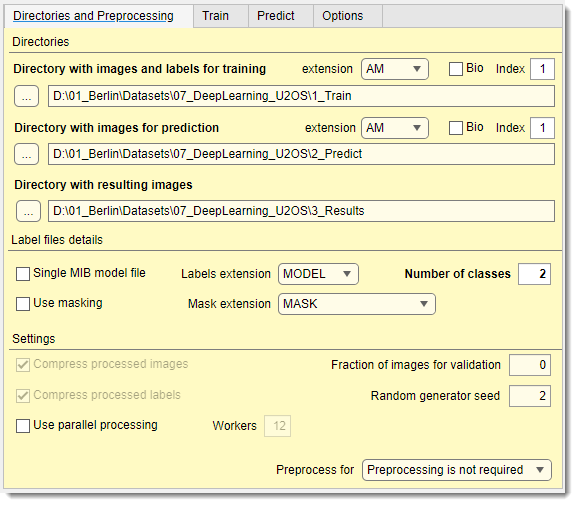
Directories
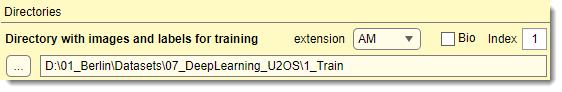
Used only for training
Select the directory containing images and models for training (named 1_Training
in directory organization schemes below).
For 2D networks, use individual 2D images; for 3D networks, use individual 3D datasets.
- : specifies the image file extension
- : toggles between standard and Bio-Formats readers. For Bio-Formats collections, use to specify
- the file index within the container
For better performance, convert Bio-Formats images to standard formats or use preprocessing (see below).
important notes considering training files
- Number of model/mask files must match the number of image files, except for 2D networks where a single
.modelfile is allowed if is checked (requires preprocessing) - For standard image format labels, specify the total number of classes (including
Exterior) in - Important: avoid numeric material names in
.modelformat; use descriptive names instead
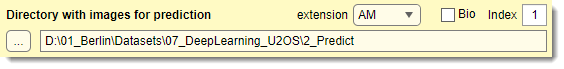
Used only for prediction
Specify the directory with images for prediction (named 2_Prediction in directory organization schemes below).
Place images in an Images subfolder (or directly in the specified folder).
Optionally, include ground truth labels in a Labels subfolder.
Info
- In preprocessing mode, images are converted and saved to
3_Results/PredictionImages. - Fround truth labels, if present, are processed to
3_Results/PredictionImages/GroundTruthLabelsfor evaluation (see Predict tab). - For 2D networks, use 2D images or 3D stacks; for 3D networks, use 3D datasets.
: specifies the image file extension
: toggles between standard and Bio-Formats readers.
For Bio-Formats collections, use to specify the file index.
Specify the main output directory where results and preprocessed images are stored. Deep MIB automatically creates subfolders:
PredictionImages: preprocessed images for predictionPredictionImages/GroundTruthLabels: ground truth labels for prediction images, if availablePredictionImages/ResultsModels: main output directory for generated labels after prediction. Combine 2D models in MIB using +during loading PredictionImages/ResultsScores: prediction scores (probability) for each material, scaled 0-255ScoreNetwork: accuracy/loss plots (if Export training plots is enabled in the Train tab) and network checkpoints (if is checked).
Scores are timestamped and overwritten with new trainingTrainImages: preprocessed training images (preprocessing mode only)TrainLabels: preprocessed training labels (preprocessing mode only)ValidationImages: preprocessed validation images (preprocessing mode only)ValidationLabels: preprocessed validation labels (preprocessing mode only)
Label file details
- : (2D networks only) uses a single
.modelfile for labels - : (2D networks only) selects the model file extension. 3D networks use MIB
.modelformat - : (TIF/PNG only) defines the number of classes, including
Exterior. Auto-updated for.modelfiles - : excludes parts of training data using masks (file count must match images). with preprocessing, requires
.maskformat; without, use for 0-value areas in labels (recommended to skip preprocessing).
Info
- With , the first predicted material is
Exterior(index 0). assign the first ground truth material to background - With preprocessing masks,
Exteriorindicates background - Masking may reduce precision due to patch inconsistency; minimize its use
- : selects mask file extension. With preprocessing, only
.maskis allowed for 3D networks; without, any format is permitted
Additional settings
- compresses preprocessed images
to
.mibImgformat (loadable in MIB or MATLAB, e.g.,res = load('img01.mibImg', '-mat');). Slows down performance - compresses preprocessed labels
to
.mibCatformat (loadable via Menu → Models → Load model). Slows performance but reduces file size significantly - enables multi-core processing, with core count set in . Speeds up preprocessing
- sets the fraction of images randomly assigned to validation (based on Random generator seed). If 0, validation is skipped
- initializes the random seed for splitting training/validation images. fixed values ensure reproducibility; If 0 uses system time for randomness
- selects the mode for the button (see schemes below)
Preprocessing of files
Preprocessing of files was originally required for most workflows, however now Deep MIB
supports unprocessed images in many cases.
In this case the Preprocess for should be set to
- to automatically split images into the training and validation sets
- after images were split into the training and validation sets
When the preprocessing step is required or recommended
Preprocessing is recommended or required when:
- Labels are in a single .MODEL file (for 2D workflows)
- Training data uses proprietary formats readable only by Bio-Formats
During preprocessing, images and models are converted to .mibImg and .mibCat formats (MATLAB-based) optimized for training and prediction.
Organization of directories
Sections below provide schemes for directories organization for several cases:
- Automatic file splitting [recommended for most cases] without file conversion, with automatic splitting of files for training and validation
- Manual file splitting without file conversion, files arranged manually into correct folders
- Conversion of files with preprocessing/file conversion, files are converted to
.mibImgand.mibCatformats and split for training and validation - Patch-wise workflow this workflow requires slightly different organization of directories
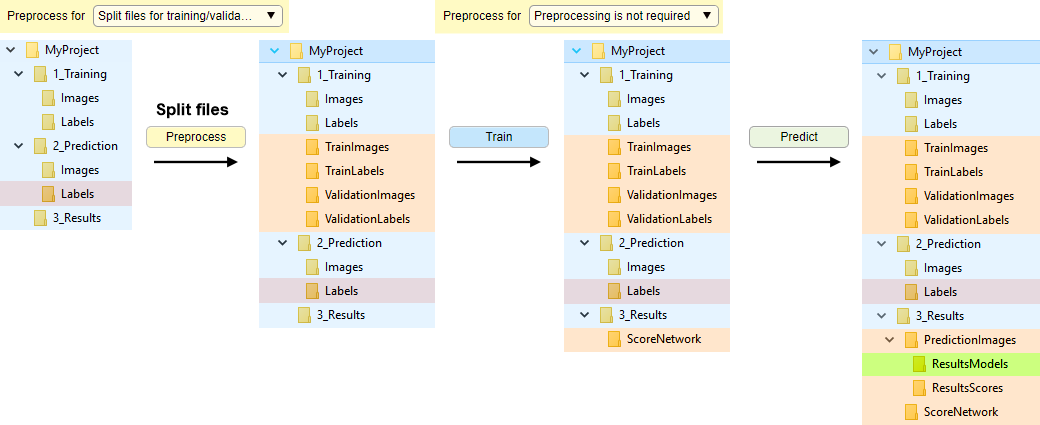
Automatic file splitting
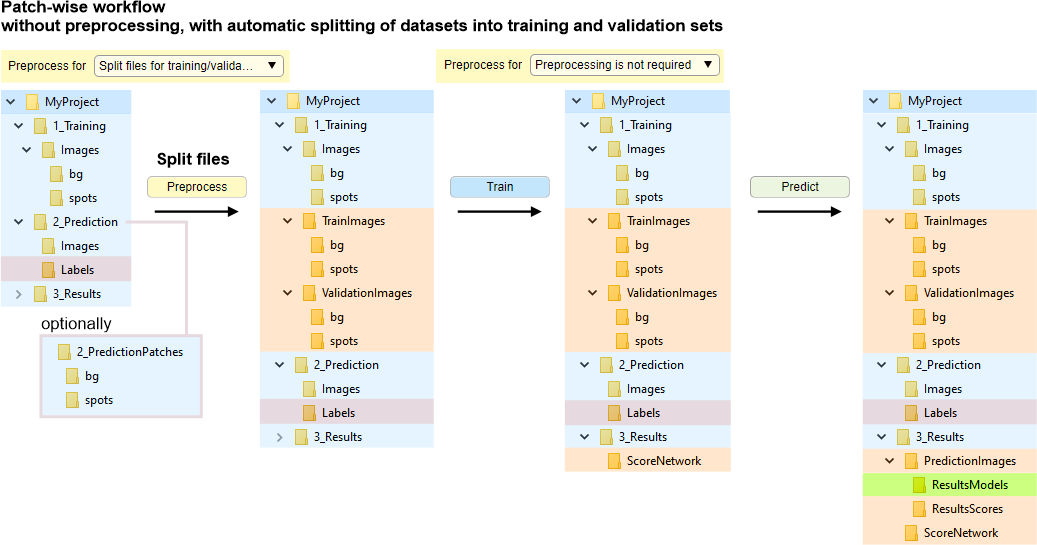
Without file conversion, with automatic splitting of datasets into training and validation sets.
Images and labels are randomly split into training and validation sets upon
clicking
when
.
Splitting depends on the
value (when "0" uses a new random seed each time).
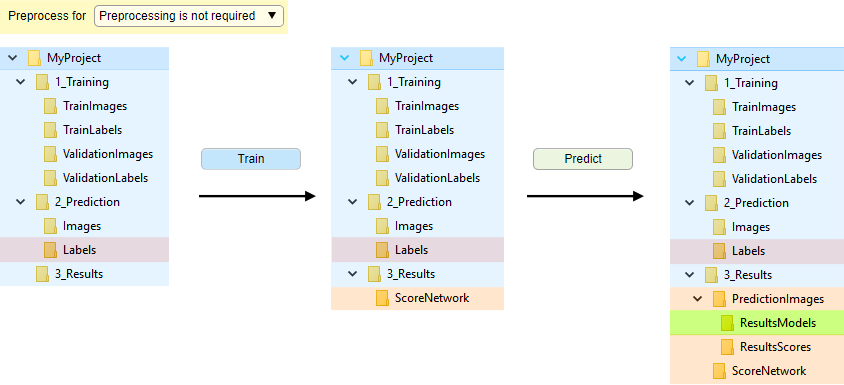
Manual file splitting
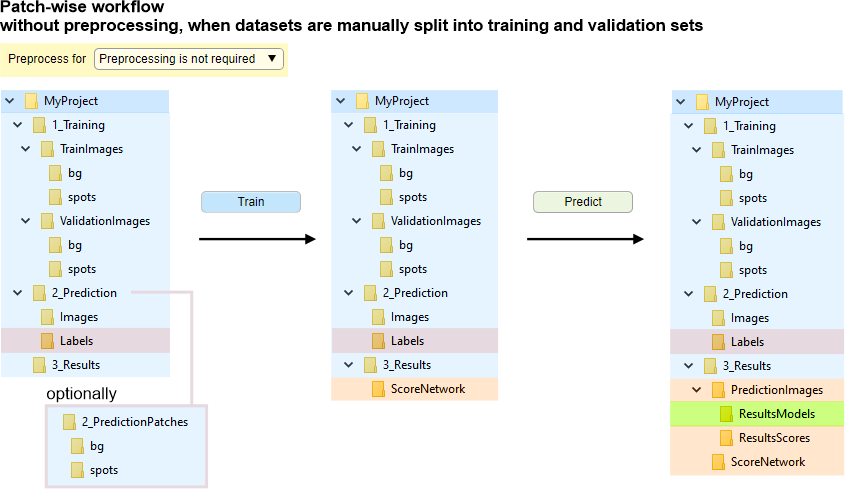
Without file conversion, when datasets are manually split into training and validation sets.
Images for training are loaded on-demand without preprocessing.
In order to proceed,
split files into TrainImages, TrainLabels, and optional ValidationImages, ValidationLabels subfolders
(see Snapshot with the directory tree below).
Automatic splitting is also available (see Automatic file splitting).
For Bio-Formats, preprocessing is recommended to improve file reading speed.
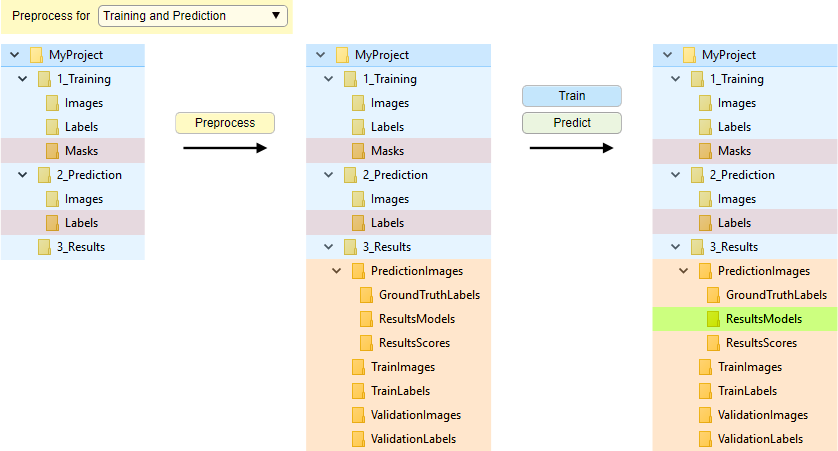
Conversion of files
Organization of directories with file conversion during preprocessing and automatic split for training and validation.
Enabled when is set to:
- Training and prediction: preprocesses for both
- Training: preprocesses only training images
- Prediction: preprocesses only prediction images
Start conversion by pressing .
Patch-wise workflow
Organization of directories for 2D patch-wise workflow. The 2D patch-wise workflow organizes training
images in Images/[ClassnameN] subfolders (e.g., bg, spots).
Number of folders should match the number of classes.
Place training images in subfolders named by class under:
- 1_Training/TrainImages: training images
- 1_Training/ValidationImages: validation images (optional)
The images can be automatic split (see the next section).
For prediction ground truth, use 2_Prediction/Images and 2_Prediction/Labels (semantic style) or 2_Prediction/[ClassnameN]
(patch-wise style).
Automatic splitting of datasets for the patch-wise segmentation into training and validation sets
Images are randomly split (based on Random generator seed) into training and
validation sets upon clicking
when .
Initially, place all images in 1_Training/Images/[ClassnameN].
Prediction ground truth follows the same options as above.
Back to MIB | User interface | DeepMIB