

Navigation
System Requirements
Contents:
|
Computer MATLAB Toolboxes |
|
| -------------------- Optional -------------------- | |
|
Bio-Formats BMxD filters DipLib library Edge Enhancing Coherence Filter Fiji Frangi Filter Imaris |
Membrane Click Tracker Omero Server Random Forest Classifier Read NRRD Segment-anything model Supervoxels/Graphcut Volume Rendering Zarr format |
Installation on Linux, contents
Requirements
- MATLAB R2022a or newer (tested on R2022a, R2022b, R2023a)
- Python 3.8, 3.9, 3.10; tested on 3.9
- List of Python versions compatible with various MATLAB releases
- Pytorch 1.10.0, (newer versions may not be compatible with Matlab installation)
- CUDA compatible GPU (CUDA 11.1) is highly recommended, CPU can also be used but it is significantly slower
Python installation
Python for SAM is installed using miniconda, the instructions below are combined from recommendations from
https://docs.anaconda.com/free/miniconda- Start terminal
- Make a new directory for miniconda installation
>> mkdir -p ~/miniconda3
- Pick a suitable miniconda installation from https://repo.anaconda.com/miniconda
Tested on Miniconda3-py39_23.11.0-2-Linux-x86_64.sh
Type the following command by replacing the corresponding link with the desired one:
>> wget https://repo.anaconda.com/miniconda/Miniconda3-py39_23.11.0-2-Linux-x86_64.sh -O ~/miniconda3/miniconda.sh
>> bash ~/miniconda3/miniconda.sh -b -u -p ~/miniconda3
>> rm -rf ~/miniconda3/miniconda.sh
- Initialize for bash shell:
>> ~/miniconda3/bin/conda init bash
- Close and reopen terminal after this operation
- Create a new environment for Python with SAM (replace python version with the installed)
>> conda create --prefix ~/miniconda3/envs/sam4mib python=3.9 - Activate the environment
>> conda activate ~/miniconda3/envs/sam4mib
Install required python packages
requirements.txt
All the following steps can be substituted by installing required packages using requirements_linux.txt
Download
(sam4mib)>> pip3 install -r requirements_linux.txt
After the installation, segment-anything can be located within "site-packages" directory of the sam4mib environment, e.g.
SAM requires python>=3.8, as well as pytorch>=1.7 and torchvision>=0.8;All the following steps can be substituted by installing required packages using requirements_linux.txt
Download
requirements_linux.txt and use the following command to install packages:After the installation, segment-anything can be located within "site-packages" directory of the sam4mib environment, e.g.
~/miniconda3/envs/sam4mib/lib/python3.9/site-packages
SAM installation was tested with Pytorch 1.10.0 and CUDA version 11.1, the newer version may not be compatible.
The list of Pytorch installation option is available from https://pytorch.org/get-started/locally
- Install pytorch:
(sam4mib)>> pip3 install torch==1.10.0+cu111 torchvision==0.11.0+cu111 torchaudio==0.10.0 -f https://download.pytorch.org/whl/torch_stable.html - The following dependencies are necessary for mask post-processing, saving masks in COCO format, the example notebooks, and exporting the model in ONNX format
(sam4mib)>> pip3 install opencv-python matplotlib onnxruntime onnx (sam4mib)>> pip3 install pycocotools -
Install "onnxruntime-gpu" to make prediction on GPU
(sam4mib)>> pip3 install onnxruntime-gpu - Forked distibution tested with MIB:
(sam4mib)>> pip3 install git+https://github.com/Ajaxels/segment-anything.git
- Distribution available on facebookresearch:
(sam4mib)>> pip3 install git+https://github.com/facebookresearch/segment-anything.git
- Define path of
python3installed in the specified environment (sam4mib):
Menu->File->Preferences->External directories->Python installation path
For example:
-~/miniconda3/envs/sam4mib/bin/python3
- Define directory to store network architectures for DeepMIB;
this location will be used to download checkpoints and onnx models. - Open SAM settings:
Segmentation panel->Segment-anything model->Settings
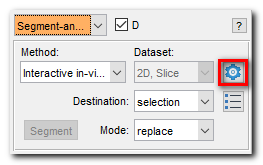
- Define location where segment-anything package was installed:
"~/miniconda3/envs/sam4mib/lib/python3.9/site-packages" - Check installation of pytorch in linux terminal:
(sam4mib)>> python -c "import torch; print(torch.__version__)" - SAM distribution has
amg.pyunderscriptssubfolder, it can be used to test the installation under python.
- Copy
amg.pyandnotebook1.png(or any other png image) from SAM distribution into directory with the downloaded checkpoint (e.g. "sam_vit_b_01ec64.pth") - Start terminal in that directory and activate sam4mib environment
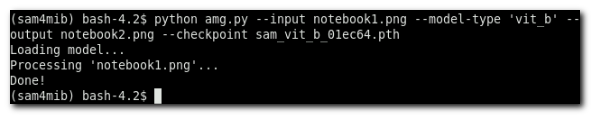
>> conda activate ~/miniconda3/envs/sam4mib - Run the following command:
(sam4mib)>> python amg.py --input notebook1.png --model-type 'vit_b' --output notebook2.png --checkpoint sam_vit_b_01ec64.pth
to load the checkpoint file and do segmentation of the image, the results are placed tonotebook2.pngsubfolder
- Copy
- Checksum test in terminal:
>> md5sum sam_vit_b_01ec64.pth - Start terminal
- List the environments installed on your system by running the command:
>> conda env list - Remove sam4mib environment:
>> conda remove --name sam4mib --all - You can also remove miniconda completely:
>> rm -rf ~/miniconda3
Install segment-anything model
There are two options, either use a forked sam distribution or the recent distribition from facebookresearch, select one of these options:
Destination!
Segment-anything will be installed to
Segment-anything will be installed to
"~/miniconda3/envs/sam4mib/lib/python3.9/site-packages"
MIB configuration
MIB configuration steps are similar to the SAM installation under
Windows with the only difference in paths.
Testing sam4mib environment
You can optionally test installation of SAM and sam4mib environment.
Remove sam4mib environment
If you do not need mib4sam environment, you can follow the following steps to uninstall it from your system.
