

Navigation
System Requirements
Contents:
|
Computer MATLAB Toolboxes |
|
| -------------------- Optional -------------------- | |
|
Bio-Formats BMxD filters DipLib library Edge Enhancing Coherence Filter Fiji Frangi Filter Imaris |
Membrane Click Tracker Omero Server Random Forest Classifier Read NRRD Segment-anything model Supervoxels/Graphcut Volume Rendering Zarr format |
Contents
General information
Zarr is a next-generation, cloud-optimized file format for storing and sharing large, multidimensional bioimaging data.
It is based on the Zarr format, enabling chunked, compressed, and scalable storage for images and metadata.
At the current state, MIB is compatible with Zarr version 2 and version 3.
- Zarr v2: the widely adopted version of Zarr, designed for storing large, chunked, compressed N-dimensional arrays, where each chunk is stored in individual file.
- Zarr v3: a newer specification aiming to improve flexibility and interoperability. It introduces a sharding mechanism for file storage optimization that groups multiple array chunks together into a single file while still allowing efficient access to individual chunks.
References
Requirements
see below for step-by-step installation instructions- MATLAB R2022b or newer (tested on R2024b)
List of Python versions compatible with various MATLAB releases - Python 3.11 or newer (tested on 3.11, version 24.5.0)
- Python environment with zarr-python packages (see below)
It is recommended to install Zarr into the same environment as segment-anything (sam4mib)!
In this case, both of those packages can be used from the same Python environment.
In this case, both of those packages can be used from the same Python environment.
- If you want to add Zarr into the segment-anything environment (sam4mib) follow to this section
- If you want to install Zarr into its own environment continue reading
------------- Installation instructions -------------
Python
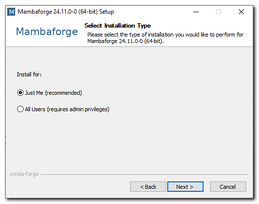
- Install Mambaforge
(tested on 24.11.0-0 with base Python 3.12, Mambaforge-24.11.0-0-Windows-x86_64.exe)
Archive of Mambaforge releases: https://github.com/conda-forge/miniforge/releases
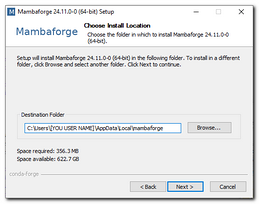
Commands below assume that Mambaforge was installed toIf you have admin account, you can install Python for all users, otherwise it is possible to install Python only for the current user.
D:\Python\Mambaforge\
Adjust the path to the actual location on Mambaforge on your system
- With Admin rights
Example of the installation directory:D:\Python\Mambaforge\
Optional note!
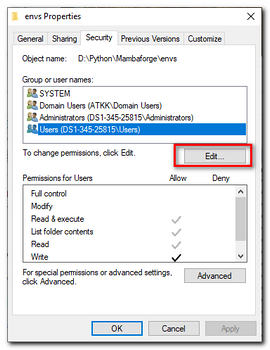
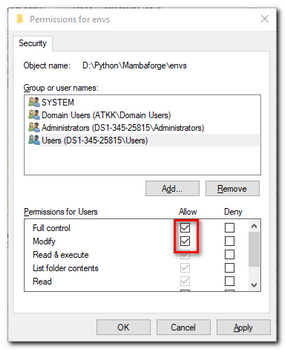
If installation was done for All Users, change permission of Mambaforge's
envsdirectory (e.g.d:\Python\Mambaforge\envs\) or the whole Mambaforge directory to be accessible for all users.
This makes things a bit more organized, otherwise the python environment will be created inC:\Users\[USERNAME]\.conda\envs\
- Without Admin rights
Example of the installation directory:C:\Users\[USERNAME]\AppData\Local\Mambaforge\
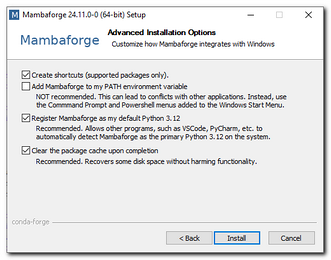
Required python packages
- Create a new environment for Python with SAM
- Start "Miniforge Prompt"
Start->Miniforge3->Miniforge prompt
 If you have a local-admin account use it to start "Anaconda Prompt" and add a new environment!
If you have a local-admin account use it to start "Anaconda Prompt" and add a new environment!
Otherwise, the installed environment may not be accessible to other users - Create a new environment, specify location for the environment and the version of Python e.g.:
>> conda create --prefix d:\Python\Mambaforge\envs\zarr4mib python=3.11
whered:\Python\Mambaforge\envs\zarr4mibdestination location for the environment.
- Start "Miniforge Prompt"
- Activate the environment:
>> activate zarr4mib
- Install Zarr for python and its dependencies
- Type this command in miniforge prompt:
(zarr4mib)>> pip install zarr
MIB configuration- Start MIB
- Open MIB preferences:
Menu->File->Preferences - Define path of
python.exeinstalled in the specified environment (zarr4mib):
External directories->Python installation path
For example:
-D:\Python\Mambaforge\envs\zarr4mib\python.exe
-C:\Users\[USERNAME]\.conda\envs\zarr4mib\python.exe
------------- How to use instructions -------------Opening OME-Zarr datasets- Switch to the virtual mode in MIB by pressing the corresponding button in the toolbar:
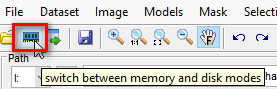
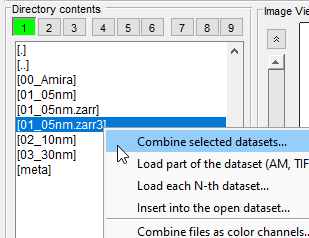
- Select Zarr folder in the Directory contents panel
- Open the dataset via
Combine selected datasets...entry of the dropdown menu:
Directory extensions!
In order to open Zarr datasets, make sure that the corresponding directory has one of the following extensions:
- *.zarr
- *.zarr2
- *.zarr3
Convert images to OME-ZarrMIB has a dedicated tool for conversion of images into OME-Zarr dataset.
Start image converter from:
MIB Menu->Plugins->File processing->Image converter
------------- Additional instructions -------------Remove zarr4mib environmentIf you do not need zarr4sam environment, you can follow the following steps to uninstall it from your system.- Start "Miniforge Prompt"
Start->Miniforge3->Miniforge prompt

- List the environments installed on your system by running the command:
>> conda env list - Remove zarr4mib environment:
>> conda remove --name zarr4mib --all
Addition of Zarr to SAM4MIB environmentIt is recommended to add Zarr to segment-anything environment (sam4mib) of MIB. In this case, the same Python environment can be used without a need for continious switch between environments.
- Start "Miniforge Prompt"
Start->Miniforge3->Miniforge prompt

- Activate the environment (if it is not yet activated):
>> activate sam4mib - Install zarr for python via pip:
(sam4mib)>> pip install zarr
- Type this command in miniforge prompt:
