|
Microscopy Image Browser 2.91
MIB
|
 |
Microscopy Image Browser 2.91
MIB
|
supporting function for mibDeepController load image function for the imagedatastore More...
Functions | |
| function imOut = | mibDeepStoreLoadImages (fn, getDataOptions) |
| supporting function for mibDeepController load image function for the imagedatastore | |
supporting function for mibDeepController load image function for the imagedatastore
| function imOut = mibDeepStoreLoadImages | ( | fn, | |
| getDataOptions ) |
supporting function for mibDeepController load image function for the imagedatastore
| fn | [string] with the full filename |
| getDataOptions | additional structure with options .mibBioformatsCheck - [logical] switch indicating use (true) or not use (false) of the BioFormats file reader .BioFormatsIndices - [numerical] index of series to be used with the BioFormats reader or index within TIF file for standard reader .Workflow - [string] used workflow, taken from obj.BatchOpt.Workflow{1} .randomCrop - [] |
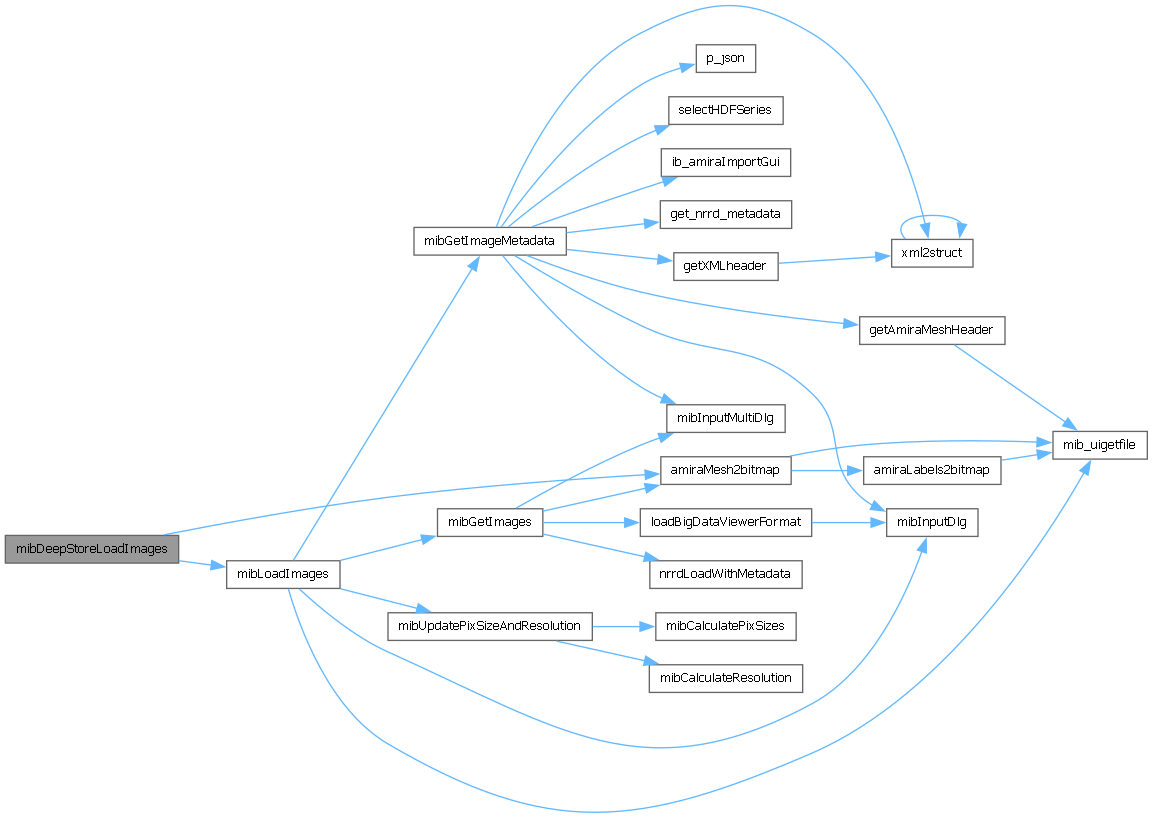
References amiraMesh2bitmap(), and mibLoadImages().
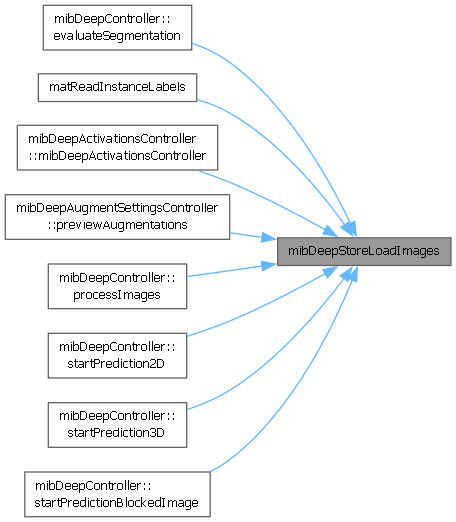
Referenced by mibDeepController.evaluateSegmentation(), matReadInstanceLabels(), mibDeepActivationsController.mibDeepActivationsController(), mibDeepAugmentSettingsController.previewAugmentations(), mibDeepController.processImages(), mibDeepController.startPrediction2D(), mibDeepController.startPrediction3D(), and mibDeepController.startPredictionBlockedImage().